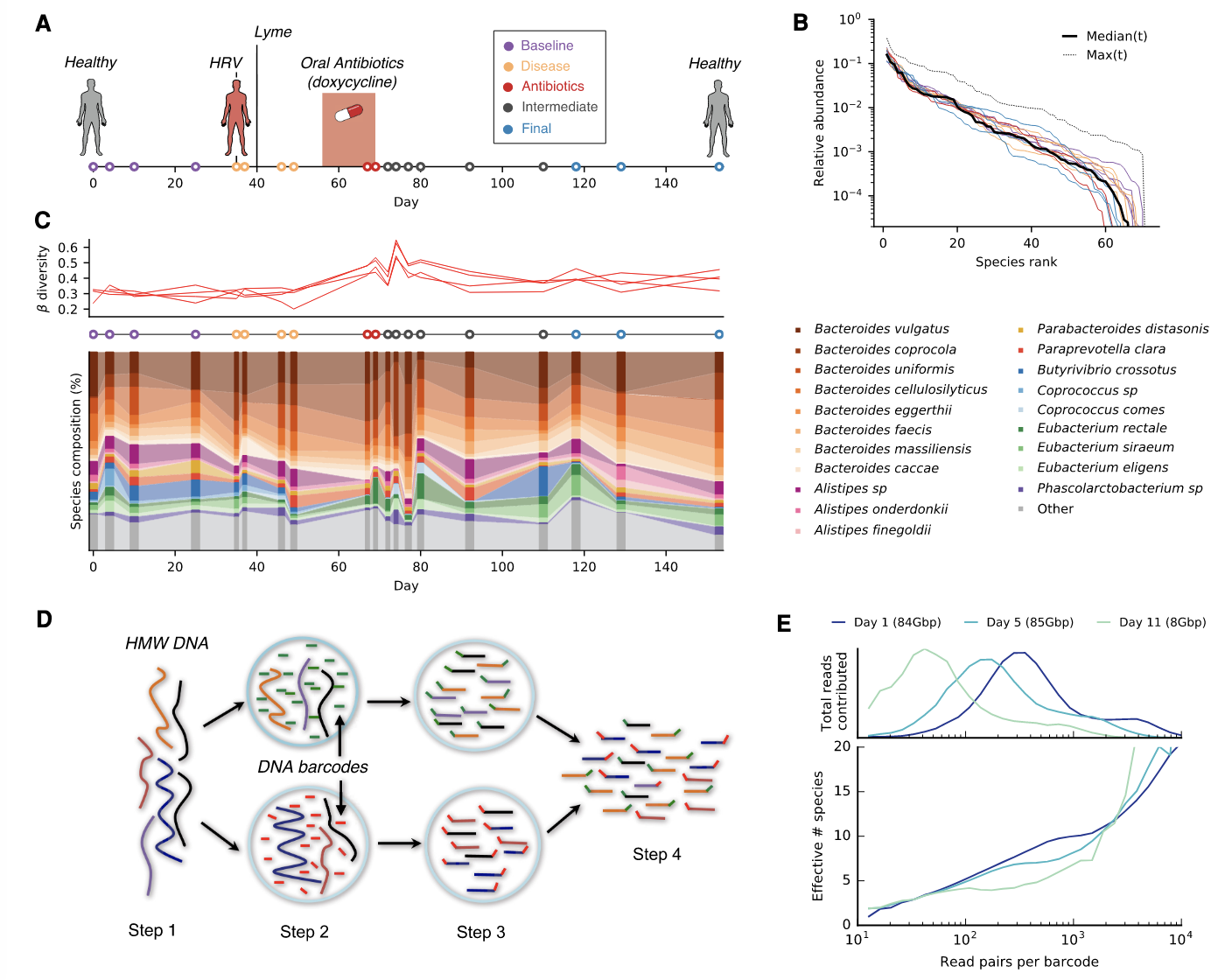
Published on the August cover of Genome Research, a team led by Dr. Morteza Roodgar used a novel method of “read cloud” DNA sequencing to obtain resolution at the subspecies level.
In this study, the team longitudinally tracked a patient’s gut microbiome for approximately 5 months, during which the patient was diagnosed with Lyme disease, and used oral antibiotics. They found changes in the gut microbiome at the subspecies level, while these changes might not be obvious at the species level.
They were able to achieve the resolution at the subspecies level because they used a novel method of “read cloud” DNA sequencing. In the read cloud method, a large fragment of DNA (also known as HMW DNA) will be located in a droplet and will be barcoded right after the DNA is sheared (broken into smaller pieces). Using these barcode labels on each smaller piece of DNA, they will find out that a small piece of DNA belongs to what original large DNA fragment before it is broken into small pieces.
Genetic changes were frequently observed in species without obvious changes in species abundance, emphasizing the importance of monitoring diversity below the species level.