Authors: Ekanath Srihari Rangan MBBS, Rahul Krishnan Pathinarupothi PhD (Amrita), Kanwaljeet J.S. Anand MBBS D.Phil and Michael P. Snyder PhD
In collaboration with Amrita University, Amritapuri, India,
Read the published paper here: https://academic.oup.com/jamiaopen/article/5/4/ooac080/6761334
In this study, we built and evaluated machine learning models for 240 different combinations of 4 vital signs: heart rate, respiratory rate, peripheral oxygen saturation, and temperature and systematically ranked these vital sign combinations with regard to 2 performance parameters: (1) area under receiver operating curve and 2) prediction times. By using second-order derived features (eg, comparing the hourly differences, cumulative differences, variance, and rate of change in the vital signs measurements), we achieved an area under the curve of 0.94, sensitivity of 82%, specificity of 85% and show that combining just heart rate with temperature predicts sepsis 6 h in advance with accuracy comparable to other sepsis predictive tools reported in literature. This lays the foundation for being able to predict progression to sepsis using noninvasive wearable sensors in limited resource settings.
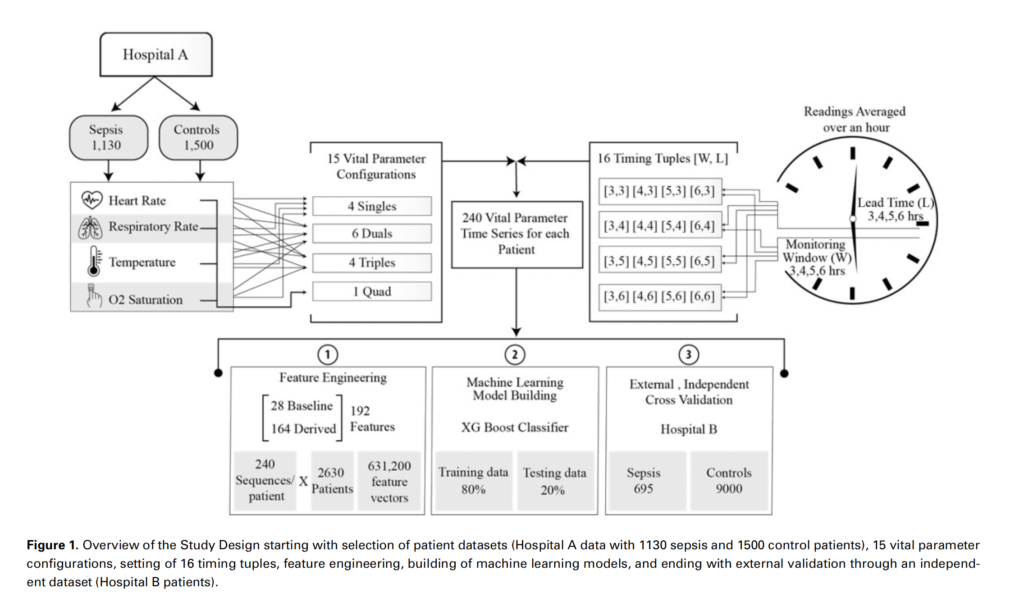
With an objective to carry out exhaustive data-driven computations for the performance of noninvasive vital signs heart rate (HR), respiratory rate (RR), peripheral oxygen saturation (SpO2), and temperature (Temp), considered both independently and in all possible combinations, for early detection of sepsis, results found that the combination of HR and Temp was found to be a minimal feature set yielding maximal predictability with area under receiver operating curve 0.94, sensitivity of 0.85, and specificity of 0.90. Whereas HR and RR each directly enhance prediction, the effects of SpO2 and Temp are significant only when combined with HR or RR. In benchmarking relative to standard methods Systemic Inflammatory Response Syndrome (SIRS), National Early Warning Score (NEWS), and quick-Sequential Organ Failure Assessment (qSOFA), Vital-SEP outperformed all 3 of them.
It can be concluded that using intensive care unit data even 2 vital signs are adequate to predict sepsis upto 6 h in advance with promising accuracy comparable to standard scoring methods and other sepsis predictive tools reported in literature. Vital-SEP can be used for fast-track prediction especially in limited resource hospital settings where laboratory based hematologic or biochemical assays may be unavailable, inaccurate, or entail clinically inordinate delays. A prospective study is essential to determine the clinical impact of the proposed sepsis prediction model and evaluate other outcomes such as mortality and duration of hospital stay.