Millions of dollars and thousands of hours are lost navigating issues surrounding healthcare big data sharing and collaboration. With more and more genomic data generated and stored in different computational platforms, genomic data analysis across multiple cloud platforms is a major ongoing challenge (especially in terms of cost and runtime).
To tackle this challenge, we built an open-source framework, Swarm, for federated cloud computation that promotes minimal data motion and facilitates crosstalk between genomic datasets stored on various cloud platforms. We demonstrate its utility via common inquiries of genomic variants across BigQuery in the Google Cloud Platform (GCP), Athena in the Amazon Web Services (AWS), Apache Presto and MySQL.
Compared to single-cloud platforms, the Swarm framework significantly reduced computational costs, run-time delays and risks of security breach and privacy violation.
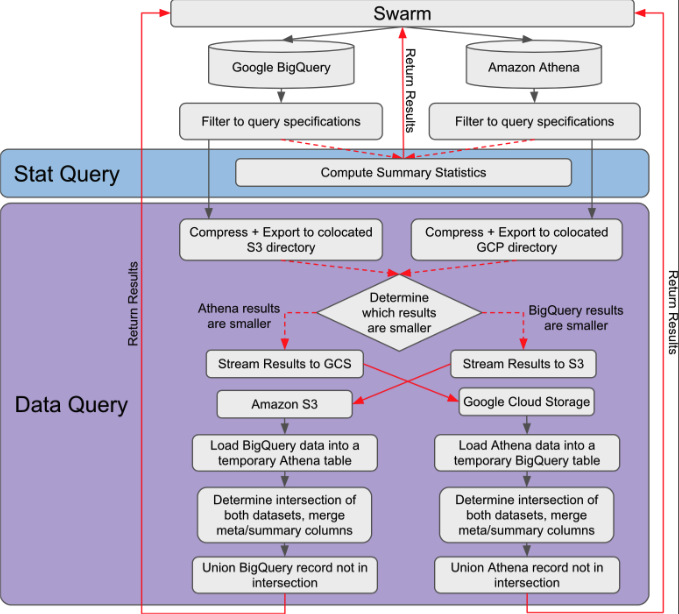
It classifies variant inquiry tasks into two main categories. “Stat Query” handles all queries that do not require data motion, and returns statistics such as counts of matched records and frequency of the alleles. “Data Query” handles queries that involve moving a set of records to another computing platform for further processing. In this figure, as an example, we illustrate the use of AWS Athena and GCP BigQuery.
Swarm facilitates easier data sharing, analysis, and collaboration within or between different organizations and institutions – saving considerable time and money. If you’re interested in learning more or utilizing Swarm, please email the team: Innovations@stanford.edu